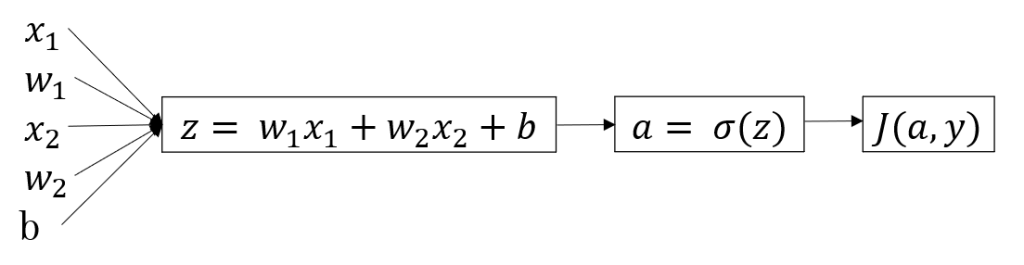In the last article, we saw how to implement logistic regression as a neural network with single neuron. In this article, we will explore how we can use Multi layered Neural Networks(multiple layers of neurons) for classifying images. We will continue exploring the same problem from last article i.e predicting the gender of the celebrity from the images.
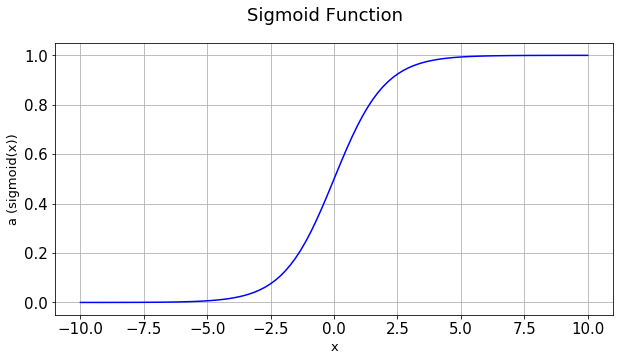
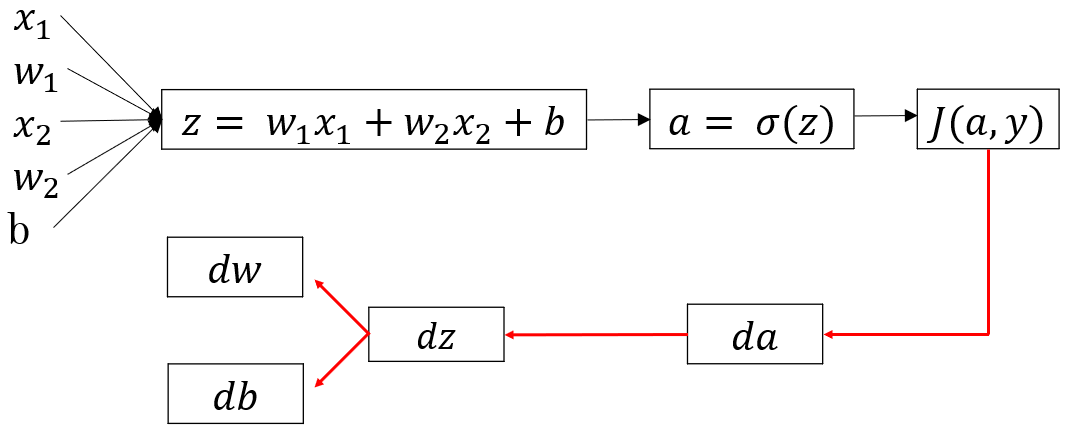
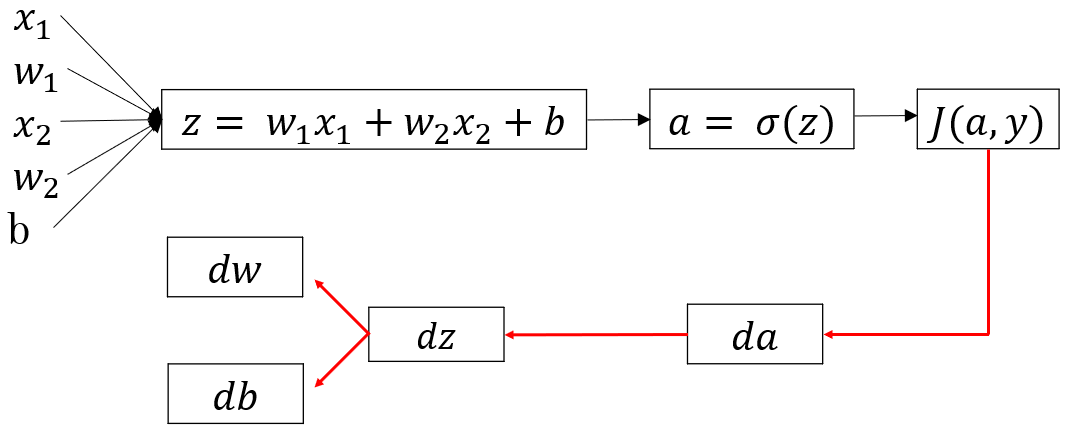
Let’s quickly recap the structure of one neuron neural network. It has mainly two components, Z and A. Z is the linear combination of inputs and A is a non-linear transformation on Z. We used sigmoid function as the non-linear transformation.
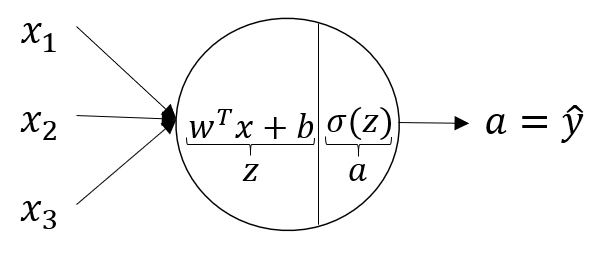
The above image shows the layout of a single neuron. x1,x2,x3 are the inputs, z is the linear function of inputs with weights W(a vector) and a is the sigmoid function of z.
Training of single neuron neural network using gradient descent involves the following steps:
1) Initialize parameters i.e W and b
2) Forward Propagation: Calculate Z and A using the initialized parameters
3) Compute cost
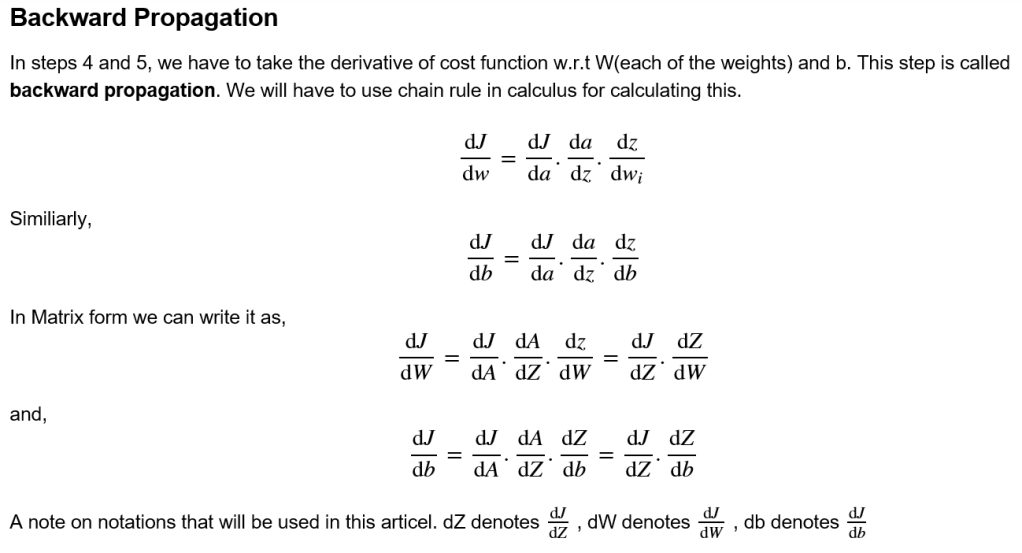
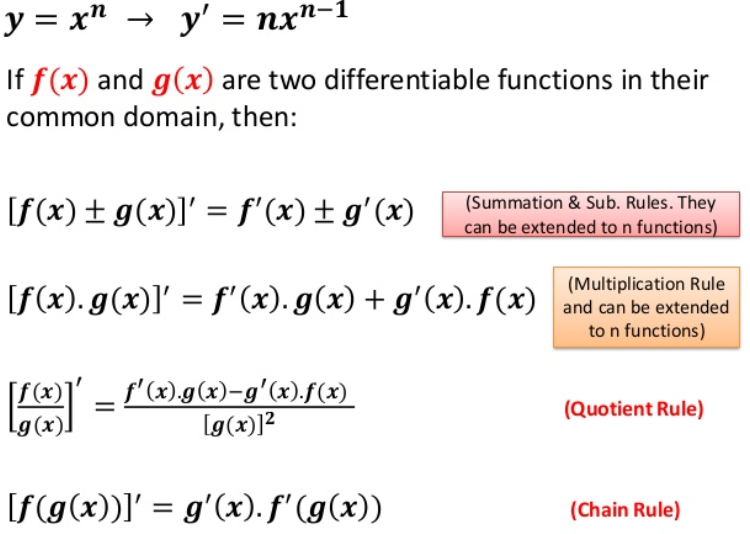
4) Backward propagation: Take gradient(derivative) of cost function with respect to W and b
5) Use the gradients to update the values of W and b
6) Repeat steps 2 to 5 for a fixed number of times
Cost is given as :
Derivatives used in backward propagation:
dA = =
dZ = =
= A-Y
dW = =
db = =
where is the transpose of X and the function to calculate a is sigmoid.
Activation Functions
Important point to remeber is that during forward propagation, we need to calculate z and a. Till now we have only used sigmoid as the function to calculate a. But we can also use other functions for a. These functions for a is called activation functions. Activation functions take z as the input and apply some form of non-linear function. Other commonly used activation functions are hyperbolic tangent, relu and leaky relu.
Tanh(hyperbolic tangent) =
Relu(Rectified Linear Units) =
Leaky relu =
Cost will remain the same for any activation function. But in back propagation, dZ will vary for different activation functions. Rest of the derivatives will be the same. The reason for change is will vary depending on the activation function.
For sigmoid, =
For tanh, = 1-
For relu, = (0, if z < 0; 1, if z >= 0)
For leaky relu, = (0.01, if z < 0; 1, if z >= 0)
From the above equations , now you can see for sigmoid function how = dZ, became A-Y.
Multi Layered Neural Networks
Architecture of Multi Layered neural networks will look something like this.
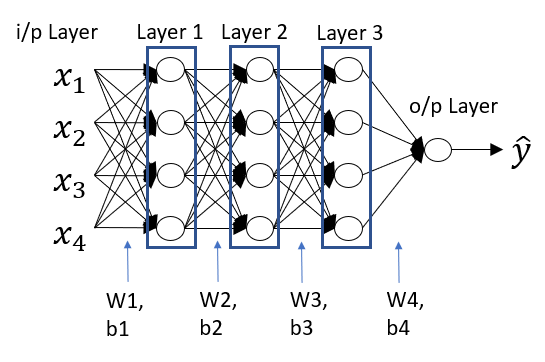
The inputs can be considered as input layers or layer 0. The last layer is the output layer. All the middle layers are called hidden layers i.e layer1,2 and 3. It’s called hidden because not much is known about what is happening inside these layers. Many people call neural networks as black box algorithm for the same reason. Understanding the working of neurons in the hidden layers is still a research area.
The calculations used for a single neuron can be expanded and generalized for multiple layers containing multiple neurons. Following figure shows the generalized calculations for each layer. It includes calculations for forward propagation and backward propagation.
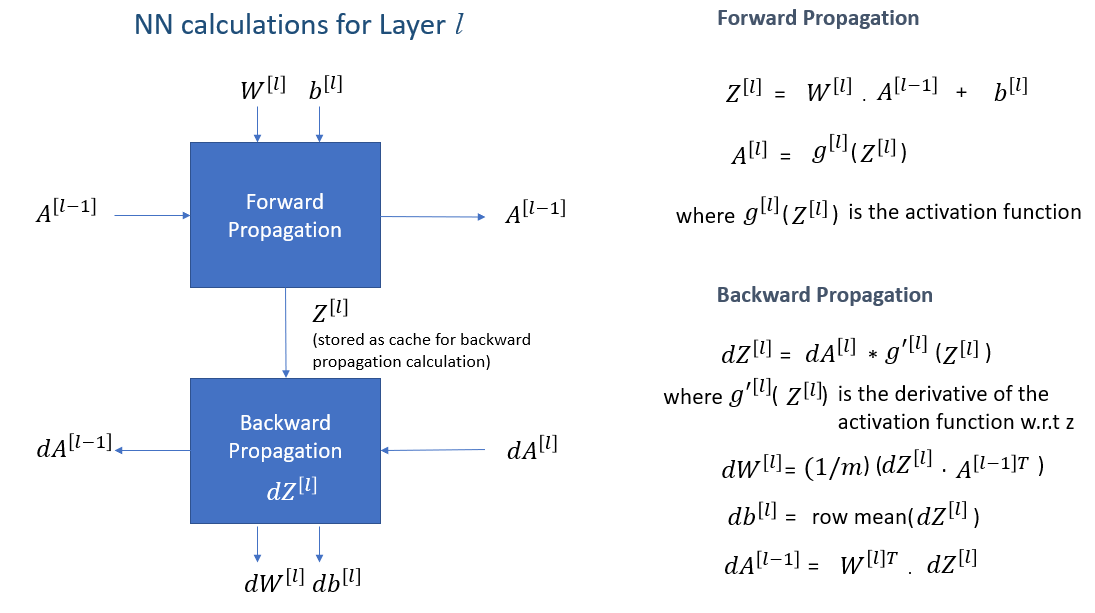
The above quations, summarize the calculations for each layer in neural networks. The superscript [] denotes the layer L. For every layer, after back propagation, weights and bias are updated as follows:
where is the learning rate.
The number of layers and number of neurons in each layer is a hyperparameter to tune. Generally, more number of layers you have, you will need less number of neurons in each layer. However, more number of layers will slow down the computations as well.
Gender Prediction from Celebrity images
Now let’s implement multi layered neural nets to solve our problem of predicting the gender of the celebrity from the image. We will use leaky relu activation for the hidden layers, three of them, and sigmoid activation for the output layer. We will continue from where we stopped in the last article.
First let’s define our activation functions, sigmoid and relu, which will take Z as the input. Sigmoid function =
and leaky relu function =
where
def sigmoid(Z):
"""
Implements the sigmoid activation in numpy
Arguments:
Z -- numpy array of any shape
Returns:
A -- output of sigmoid(z), same shape as Z
cache -- returns Z as well, useful during backpropagation
"""
A = 1/(1+np.exp(-Z))
cache = Z
return A, cache
def relu(Z):
"""
Implement the RELU function.
Arguments:
Z -- Output of the linear layer, of any shape
Returns:
A -- Post-activation parameter, of the same shape as Z
cache -- a python dictionary containing "A" ; stored for computing the backward pass efficiently
"""
A = np.maximum(0.01*Z,Z)
assert(A.shape == Z.shape)
cache = Z
return A, cache
Now let’s define functions for backpropagation of activation function. i.e for sigmoid and relu function.
For sigmoid, =
For leaky relu, = (0, if z < 0; 1, if z >= 0)
We will pass dA and Z(stored as cache) as inputs to the function.
def relu_backward(dA, cache):
"""
Implement the backward propagation for a single RELU unit.
Arguments:
dA -- post-activation gradient, of any shape
cache -- 'Z' where we store for computing backward propagation efficiently
Returns:
dZ -- Gradient of the cost with respect to Z
"""
Z = cache
dZ = np.array(dA, copy=True) # just converting dz to a correct object.
# When z <= 0, you should set dz to 0 as well.
dZ[Z < 0] = dZ[Z < 0]*0.01
assert (dZ.shape == Z.shape)
return dZ
def sigmoid_backward(dA, cache):
"""
Implement the backward propagation for a single SIGMOID unit.
Arguments:
dA -- post-activation gradient, of any shape
cache -- 'Z' where we store for computing backward propagation efficiently
Returns:
dZ -- Gradient of the cost with respect to Z
"""
Z = cache
s = 1/(1+np.exp(-Z))
dZ = dA * s * (1-s)
assert (dZ.shape == Z.shape)
return dZ
Now let us write a function to intialize the parameters, weights and bias for every layer. Unlike the single neuron case, we have to intialize weights for each layers separately. Weights for every layer will have the shape[no. of neurons in that layer, no. of neurons in the previous layer]. Bias for each layer will have the shape [no. of neurons in that layer,1].
Also, we will initialize weights from a standard normal distribution and multiply by . This is called he intialization. Generally this intialization works well. Biases can be intialized as zeros.
Random intialization of weights are better since it will allow different neurons to learn different aspects of the image. Also, the reason for drawing random numbers from a standard normal distribution(mean=0,std.dev=1) is that this will help in avoiding a problem called exploding or vanishing gradients. I am not going into the details but vanishing gradient in a nutshell means, during backpropagation of a deep neural network, the gradients of the intial layers become so small that they practically cease to update, SImiliarly, Exploding gradients means gradients of intial layers quickly reach a higher number or infinity stopping further learning. Vanishing gradients were one of the reasons why deep learning didn’t become popular until 2006. I didn’t mention earlier why we are using leaky relu activation function for the hidden layers. Leaky relus are better in handling vanishing or exploding gradients.
def initialize_parameters_deep(layer_dims):
"""
Arguments:
layer_dims -- python array (list) containing the dimensions of each layer in our network
Returns:
parameters -- python dictionary containing your parameters "W1", "b1", ..., "WL", "bL":
Wl -- weight matrix of shape (layer_dims[l], layer_dims[l-1])
bl -- bias vector of shape (layer_dims[l], 1)
"""
np.random.seed(1)
parameters = {}
L = len(layer_dims) # number of layers in the network
for l in range(1, L):
parameters['W' + str(l)] = np.random.randn(layer_dims[l], layer_dims[l-1]) * np.sqrt(2/layer_dims[l-1]) #*0.01
parameters['b' + str(l)] = np.zeros((layer_dims[l], 1))
assert(parameters['W' + str(l)].shape == (layer_dims[l], layer_dims[l-1]))
assert(parameters['b' + str(l)].shape == (layer_dims[l], 1))
return parameters
Let’s get to the linear forward function. This function will calculate the linear function , for one layer (for all neurons in the layer) at a time. It will take A from previous layer and W(weight) and b(bias) of current layer as input.
def linear_forward(A, W, b):
"""
Implement the linear part of a layer's forward propagation.
Arguments:
A -- activations from previous layer (or input data): (size of previous layer, number of examples)
W -- weights matrix: numpy array of shape (size of current layer, size of previous layer)
b -- bias vector, numpy array of shape (size of the current layer, 1)
Returns:
Z -- the input of the activation function, also called pre-activation parameter
cache -- a python dictionary containing "A", "W" and "b" ; stored for computing the backward pass efficiently
"""
Z = W.dot(A) + b
assert(Z.shape == (W.shape[0], A.shape[1]))
cache = (A, W, b)
return Z, cache
Let’s write a function to calculate the derivative values we need in each layer. We already wrote function to calculated Z. In this function we will calculate dW, dB and dA_prev for one layer at a time.
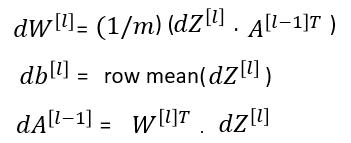
def linear_backward(dZ, cache):
"""
Implement the linear portion of backward propagation for a single layer (layer l)
Arguments:
dZ -- Gradient of the cost with respect to the linear output (of current layer l)
cache -- tuple of values (A_prev, W, b) coming from the forward propagation in the current layer
Returns:
dA_prev -- Gradient of the cost with respect to the activation (of the previous layer l-1), same shape as A_prev
dW -- Gradient of the cost with respect to W (current layer l), same shape as W
db -- Gradient of the cost with respect to b (current layer l), same shape as b
"""
A_prev, W, b = cache
m = A_prev.shape[1]
dW = 1./m * np.dot(dZ,A_prev.T)
db = 1./m * np.sum(dZ, axis = 1, keepdims = True)
dA_prev = np.dot(W.T,dZ)
assert (dA_prev.shape == A_prev.shape)
assert (dW.shape == W.shape)
assert (db.shape == b.shape)
return dA_prev, dW, db
Let’s define a common activation function for forward propagation and backward propagation, which will take activation as one of the input. Depending on the kind of value is passed for activation, the function will do the calculations for sigmoid or leaky relu.
def linear_activation_forward(A_prev, W, b, activation):
"""
Implement the forward propagation for the LINEAR->ACTIVATION layer
Arguments:
A_prev -- activations from previous layer (or input data): (size of previous layer, number of examples)
W -- weights matrix: numpy array of shape (size of current layer, size of previous layer)
b -- bias vector, numpy array of shape (size of the current layer, 1)
activation -- the activation to be used in this layer, stored as a text string: "sigmoid" or "relu"
Returns:
A -- the output of the activation function, also called the post-activation value
cache -- a python dictionary containing "linear_cache" and "activation_cache";
stored for computing the backward pass efficiently
"""
if activation == "sigmoid":
# Inputs: "A_prev, W, b". Outputs: "A, activation_cache".
Z, linear_cache = linear_forward(A_prev, W, b)
A, activation_cache = sigmoid(Z)
elif activation == "relu":
# Inputs: "A_prev, W, b". Outputs: "A, activation_cache".
Z, linear_cache = linear_forward(A_prev, W, b)
A, activation_cache = relu(Z)
assert (A.shape == (W.shape[0], A_prev.shape[1]))
cache = (linear_cache, activation_cache)
return A, cache
def linear_activation_backward(dA, cache, activation):
"""
Implement the backward propagation for the LINEAR->ACTIVATION layer.
Arguments:
dA -- post-activation gradient for current layer l
cache -- tuple of values (linear_cache, activation_cache) we store for computing backward propagation efficiently
activation -- the activation to be used in this layer, stored as a text string: "sigmoid" or "relu"
Returns:
dA_prev -- Gradient of the cost with respect to the activation (of the previous layer l-1), same shape as A_prev
dW -- Gradient of the cost with respect to W (current layer l), same shape as W
db -- Gradient of the cost with respect to b (current layer l), same shape as b
"""
linear_cache, activation_cache = cache
if activation == "relu":
dZ = relu_backward(dA, activation_cache)
dA_prev, dW, db = linear_backward(dZ, linear_cache)
elif activation == "sigmoid":
dZ = sigmoid_backward(dA, activation_cache)
dA_prev, dW, db = linear_backward(dZ, linear_cache)
return dA_prev, dW, db
Now let’s define a function to create any number of layers of neurons from the functions we created. Since we have generalized calculations for each layer, we can add any number of layers. This function will do the forward propagation calcuations iteratively for each layer, starting from the input layer and ending with the output layer.
def L_model_forward(X, parameters):
"""
Implement forward propagation for the [LINEAR->RELU]*(L-1)->LINEAR->SIGMOID computation
Arguments:
X -- data, numpy array of shape (input size, number of examples)
parameters -- output of initialize_parameters_deep()
Returns:
AL -- last post-activation value
caches -- list of caches containing:
every cache of linear_relu_forward() (there are L-1 of them, indexed from 0 to L-2)
the cache of linear_sigmoid_forward() (there is one, indexed L-1)
"""
caches = []
A = X
L = len(parameters) // 2 # number of layers in the neural network. Every layer has a weight and bias, hence the length of the parameter will be double the layers.
# Implement [LINEAR -> RELU]*(L-1). Add "cache" to the "caches" list.
for l in range(1, L):
A_prev = A
A, cache = linear_activation_forward(A_prev, parameters['W' + str(l)], parameters['b' + str(l)], activation = "relu")
caches.append(cache)
# Implement LINEAR -> SIGMOID. Add "cache" to the "caches" list.
AL, cache = linear_activation_forward(A, parameters['W' + str(L)], parameters['b' + str(L)], activation = "sigmoid")
caches.append(cache)
assert(AL.shape == (1,X.shape[1]))
return AL, caches
Cost is calculated after the prediction from output layer is obtained. The formula for cost is same as in the last article(the same cost works for all binomial classification problems, although it can be generalized to multi-class classification problem).
def compute_cost(AL, Y):
"""
Implement the cost function defined by equation (7).
Arguments:
AL -- probability vector corresponding to your label predictions, shape (1, number of examples)
Y -- true "label" vector (for example: containing 0 if non-cat, 1 if cat), shape (1, number of examples)
Returns:
cost -- cross-entropy cost
"""
m = Y.shape[1]
# Compute loss from aL and y.
cost = (1./m) * (-np.dot(Y,np.log(AL).T) - np.dot(1-Y, np.log(1-AL).T))
cost = np.squeeze(cost) # To make sure your cost's shape is what we expect (e.g. this turns [[17]] into 17).
assert(cost.shape == ())
return cost
Now let’s define a function to do the backprop calculations for all the layers we created during forward propagation. Each layer calculation is done iteratively, starting from the output layer and ending with the input layer.
def L_model_backward(AL, Y, caches):
"""
Implement the backward propagation for the [LINEAR->RELU] * (L-1) -> LINEAR -> SIGMOID group
Arguments:
AL -- probability vector, output of the forward propagation (L_model_forward())
Y -- true "label" vector (containing 0 if non-cat, 1 if cat)
caches -- list of caches containing:
every cache of linear_activation_forward() with "relu" (there are (L-1) or them, indexes from 0 to L-2)
the cache of linear_activation_forward() with "sigmoid" (there is one, index L-1)
Returns:
grads -- A dictionary with the gradients
grads["dA" + str(l)] = ...
grads["dW" + str(l)] = ...
grads["db" + str(l)] = ...
"""
grads = {}
L = len(caches) # the number of layers
m = AL.shape[1]
Y = Y.reshape(AL.shape) # after this line, Y is the same shape as AL
# Initializing the backpropagation
dAL = - (np.divide(Y, AL) - np.divide(1 - Y, 1 - AL))
# Lth layer (SIGMOID -> LINEAR) gradients. Inputs: "AL, Y, caches". Outputs: "grads["dAL"], grads["dWL"], grads["dbL"]
current_cache = caches[L-1]
grads["dA" + str(L)], grads["dW" + str(L)], grads["db" + str(L)] = linear_activation_backward(dAL, current_cache, activation = "sigmoid")
for l in reversed(range(L-1)):
# lth layer: (RELU -> LINEAR) gradients.
current_cache = caches[l]
dA_prev_temp, dW_temp, db_temp = linear_activation_backward(grads["dA" + str(l + 2)], current_cache, activation = "relu")
grads["dA" + str(l + 1)] = dA_prev_temp
grads["dW" + str(l + 1)] = dW_temp
grads["db" + str(l + 1)] = db_temp
return grads
Let’s write a function to update the parameters using the calculated gradients from backprop. This function can actually be combined in the backprop step. However, to enhance understanding let’s write it separately.
def update_parameters(parameters, grads, learning_rate):
"""
Update parameters using gradient descent
Arguments:
parameters -- python dictionary containing your parameters
grads -- python dictionary containing your gradients, output of L_model_backward
Returns:
parameters -- python dictionary containing your updated parameters
parameters["W" + str(l)] = ...
parameters["b" + str(l)] = ...
"""
L = len(parameters) // 2 # number of layers in the neural network
# Update rule for each parameter. Use a for loop.
for l in range(L):
parameters["W" + str(l+1)] = parameters["W" + str(l+1)] - learning_rate * grads["dW" + str(l+1)]
parameters["b" + str(l+1)] = parameters["b" + str(l+1)] - learning_rate * grads["db" + str(l+1)]
return parameters
Time to put everything together and create a single function for Multi layered neural network. Just to remind, the steps involved are:
1) Initialize parameters i.e W and b
2) Forward Propagation: Calculate Z and A using the initialized parameters
3) Compute cost
4) Backward propagation: Take gradient(derivative) of cost function with respect to W and b
5) Use the gradients to update the values of W and b
6) Repeat steps 2 to 5 for a fixed number of times(num_iterations)
def L_layer_model(X, Y, layers_dims, learning_rate = 0.0075, num_iterations = 3000, print_cost=False):
"""
Implements a L-layer neural network: [LINEAR->RELU]*(L-1)->LINEAR->SIGMOID.
Arguments:
X -- data, numpy array of shape (number of examples, num_px * num_px * 3)
Y -- true "label" vector (containing 0 if cat, 1 if non-cat), of shape (1, number of examples)
layers_dims -- list containing the input size and each layer size, of length (number of layers + 1).
learning_rate -- learning rate of the gradient descent update rule
num_iterations -- number of iterations of the optimization loop
print_cost -- if True, it prints the cost every 100 steps
Returns:
parameters -- parameters learnt by the model. They can then be used to predict.
"""
np.random.seed(1)
costs = [] # keep track of cost
# Parameters initialization.
### START CODE HERE ###
parameters = initialize_parameters_deep(layers_dims)
### END CODE HERE ###
# Loop (gradient descent)
for i in range(0, num_iterations):
# Forward propagation: [LINEAR -> RELU]*(L-1) -> LINEAR -> SIGMOID.
### START CODE HERE ### (≈ 1 line of code)
AL, caches = L_model_forward(X, parameters)
### END CODE HERE ###
# Compute cost.
### START CODE HERE ### (≈ 1 line of code)
cost = compute_cost(AL, Y)
### END CODE HERE ###
# Backward propagation.
### START CODE HERE ### (≈ 1 line of code)
grads = L_model_backward(AL, Y, caches)
### END CODE HERE ###
# Update parameters.
### START CODE HERE ### (≈ 1 line of code)
parameters = update_parameters(parameters, grads, learning_rate)
### END CODE HERE ###
# Print the cost every 100 training example
if print_cost and i % 100 == 0:
print ("Cost after iteration %i: %f" %(i, cost))
if print_cost and i % 100 == 0:
costs.append(cost)
# plot the cost
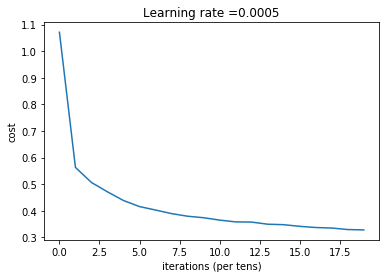
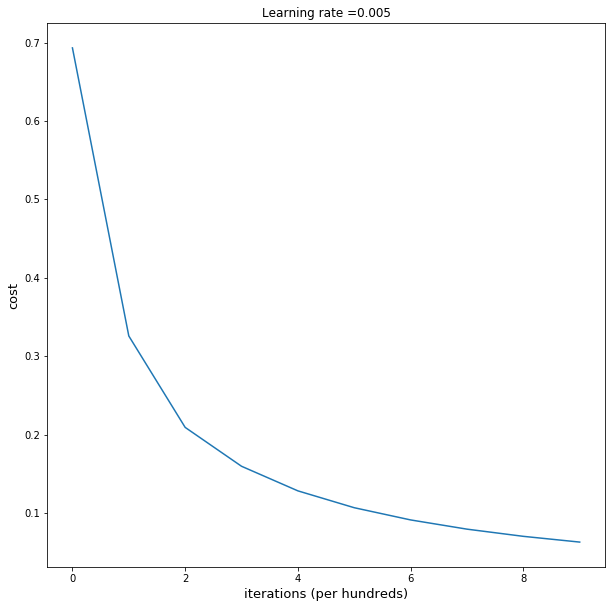
plt.plot(np.squeeze(costs))
plt.ylabel('cost')
plt.xlabel('iterations (per tens)')
plt.title("Learning rate =" + str(learning_rate))
plt.show()
return parameters
Now let us design the number of layers an neurons we need for our model. The size of the input is 12288(flattened pixel size for each image) and size of output layer is 1. We can decide on any number of input layers by adding more values in between. The number in between decides the number of neurons in that layer.
### CONSTANTS ###
layers_dims = [12288, 20, 7,5, 1] # 5-layer model, 3 hidden layers, 1 input and 1 output layer
Let us run the model on our training dataset.
parameters = L_layer_model(train_x, y_train, layers_dims,learning_rate = 0.0005, num_iterations = 2000, print_cost = True)